Models
Navigation
Below is displayed the model view of the selected project. Model view is shown in the form of the model overview page for the currently selected model. The central feature of the model view is the model scheme that shows individual model components of selected model. The navigation panel on the left allows you to browse the biological structure of the model. Manipulation with the navigation panel is realized by unfolding the items in the navigation tree and clicking on a requested system level.
Annotations Tab
All the annotation terms relevant for the currently focused level of the project are displayed on the Annotation Tab below the scheme. Individual annotation data can be unfolded by clicking on the requested annotation item header.
Components Tab
The Components Tab displays all the model species (state variables). More information for particular components are accessible after clicking on the requested component header.
Reactions Tab
Reactions Tab contains information regarding the modeled reactions. After clicking on the particular reaction header, the reacting components and relevant kinetic parameters are displayed.
Parameters Tab
All quantitative parameters are managed under Parameters Tab. Constants are separated from assigned quantities.
Simulation Tab
Simulation and SBML export are available by clicking on appropriate buttons at the bottom of the tab. All relevant settings of initial conditions, parameters, options and datasets are listed in respective folders.
Analysis Tab
Conservation analysis, modes analysis and matrix analysis are available by clicking on appropriate buttons.
Experiments Tab
Experiments tab contains list of all experiments related to selected model.
Müller et al. 2019
Towards a quantitative assessment of inorganic carbon cycling in photosynthetic microorganisms
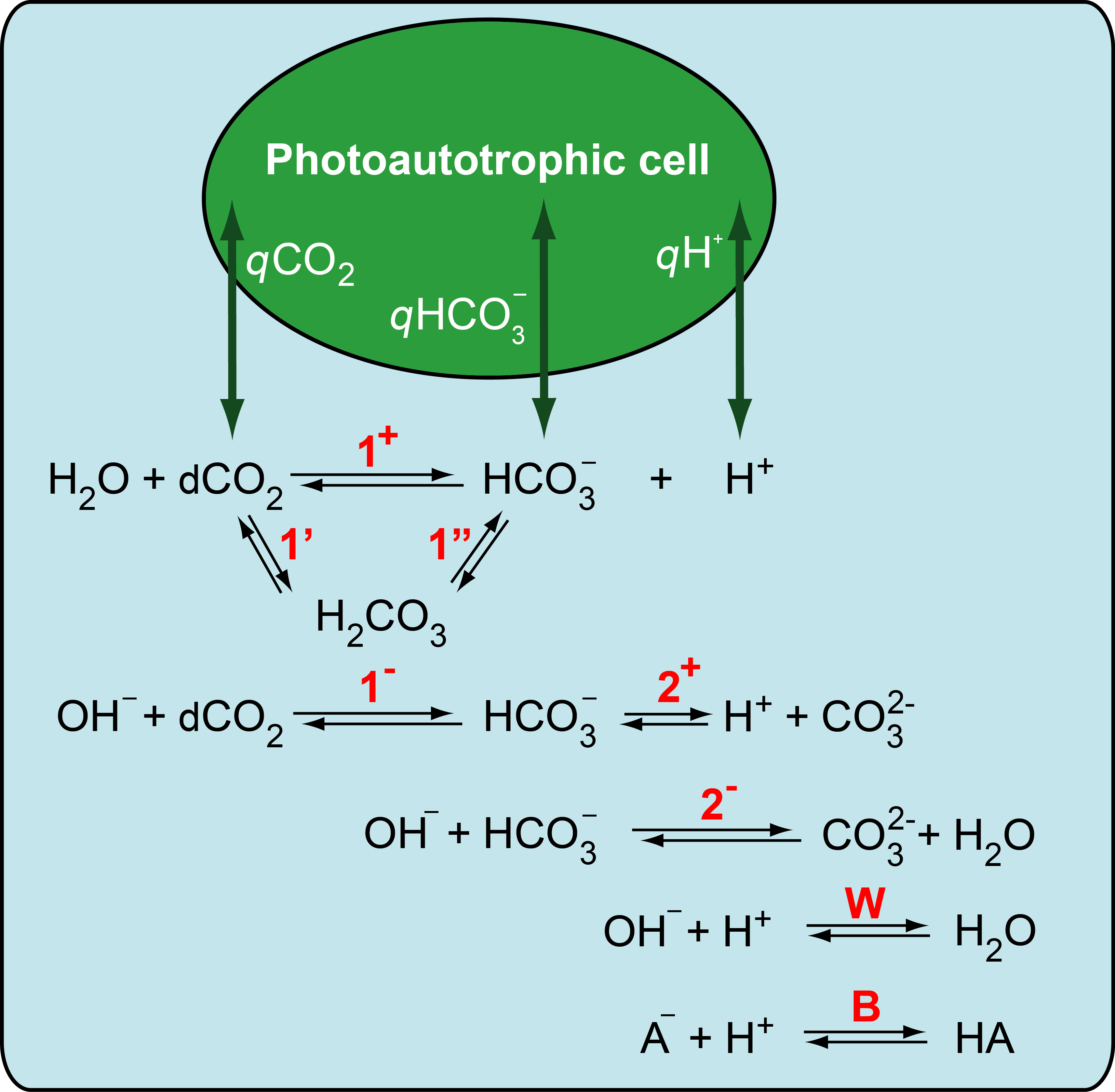
Complete version of the model will be available soon on newly rebuilt version of the webpage.
Photosynthetic organisms developed various strategies to mitigate high light stress. For instance, aquatic organisms are able to spend excessive energy by exchanging dissolvedcarbon dioxide (dCO2) and bicarbonate (HCO32-) with the environment. Simultaneous uptake and excretion of the two carbon species is referred to as inorganic carbon cycling (ICC). Often, ICC is indicated by displacements of the extracellular dCO2 signal from the equilibrium value after changing the light conditions. In this work, we additionally use (i) the extracellular pH signal, which requires non- or weakly-buffered medium, and (ii) a dynamic model of carbonate chemistry in the aquatic environment in order to detect and quantitatively describe ICC. Based on simulations and experiments in precisely controlled photobioreactors,we show that the magnitude of the observed dCO2 displacement crucially depends on extracellular pH level and buffer concentration. Moreover, we find that the dCO2 displacement can also be caused by simultaneous uptake of both dCO2 and HCO32- (no ICC). In a next step, the dynamic model of carbonate chemistry allows for a quantitative assessment of cellular dCO2 , HCO32- , and H+ exchange rates from the measured dCO2 and pH signals. Limitations of the method are discussed.
model: Müller et al. 2019
Müller S, Zavřel T, Červený J (2019) Towards a quantitative assessment of inorganic carbon cycling in photosynthetic microorganisms. Eng Life Sci:1-13
publication: Müller et al. 2019
Simulation type: fixed
Simulation type: reaction
Simulation type: reaction
Simulation type: reaction
Simulation type: reaction
Simulation type: reaction
Simulation type: reaction
Simulation type: reaction
Constant quantities
Assigned quantities
M. Trojak, D. Safranek, J. Hrabec, J. Salagovic, F. Romanovska, J. Cerveny: E-Cyanobacterium.org: A Web-Based Platform for Systems Biology of Cyanobacteria. In: Computational Methods in Systems Biology, CMSB 2016, Vol. 9859 of LNCS, pp. 316-322. Springer, 2016. DOI
