This page displays basic information about selected experiment.
In case of series of experiments, these can be found below in List of experiments with links
| Series name: | Figure 6 Experimental data of Synechocystis sp. PCC 6803 cultivated in non-buffe |
|---|---|
| Summary: | measurements of dCO2 (full blue lines) and pH signals (full black lines) signals (left panels) for initial pH 6.5 (panel A), 7.3 (panel C), 7.7 (panel E), and 8.1 (panel G). Four biological replicates with qualitatively identical results for each condition were collected; data from one representative experiment are shown. Identification of the exchange rates qCO2 (dotted blue lines, right panels) and qHCO3- (dotted black lines, right panels), identified from the measured data involving data differentiation and the assumption of qH+ = qHCO3- is represented in right panels for initial pH 6.5 (panel B), 7.3 (panel D), 7.7 (panel F), and 8.1 (panel H). Positive values in the right panels indicate dCO2 or HCO3- excretion, negative values indicate dCO2 or HCO3- uptake. For further experimental details, see Section 2. |
| Inserted: | 2019-10-22 07:22:12 |
| Last update: | 2019-10-22 10:32:52 |
| Number of experiments: | 4 |
List of experiments
| Name | Started | Inserted | |
|---|---|---|---|
| Panel B | 2019-10-22 08:31:46 | ||

|
Panel A | 2019-10-22 07:37:55 | |
| Panel C | 2019-10-22 08:39:24 | ||
| Panel D | 2019-10-22 08:43:36 |
| Name: | Panel A |
|---|---|
| Device Type: | Photobioreactor-041 |
| Device Name: | DoAB-PBR01 |
| Device ID: | 72700001 |
| Name: | Panel A: dCO2 |
|---|---|
| Device Type: | PSI MS GAS-100 |
| Device Name: | MIMS Prototype |
| Device ID: | MIMS-001 |
| Code: | _ph |
|---|---|
| Number of values: | 613 |
| Related to: |
Müller et al. 2019: [H.+]
|
| Code: | _dco2 |
|---|---|
| Number of values: | 274 |
| Related to: |
Müller et al. 2019: [CO2]
|
| From: | Panel A: dCO2 |
This page displays graph presenting the requested experiment data. You can export data in on of the available formats or show chart.
Individual variables can be switched on/off by clicking on the respective legend. Using corresponding dropdown list it is possible to select desired Graphset of variables or Dataset. Button on the top left side allows to print graph or download it as image in corresponding format. The graph can be zoomed selecting the area that you want to zoom by mouse. Clicking button Reset zoom will show whole graph back. It is also possible to Change axis type or Fit to scale by clicking corresponding buttons. Simulation data and Simulation source could be downloaded by clicking at desired file format at the bottom of the page.
Show chart
Towards a quantitative assessment of inorganic carbon cycling in photosynthetic microorganisms
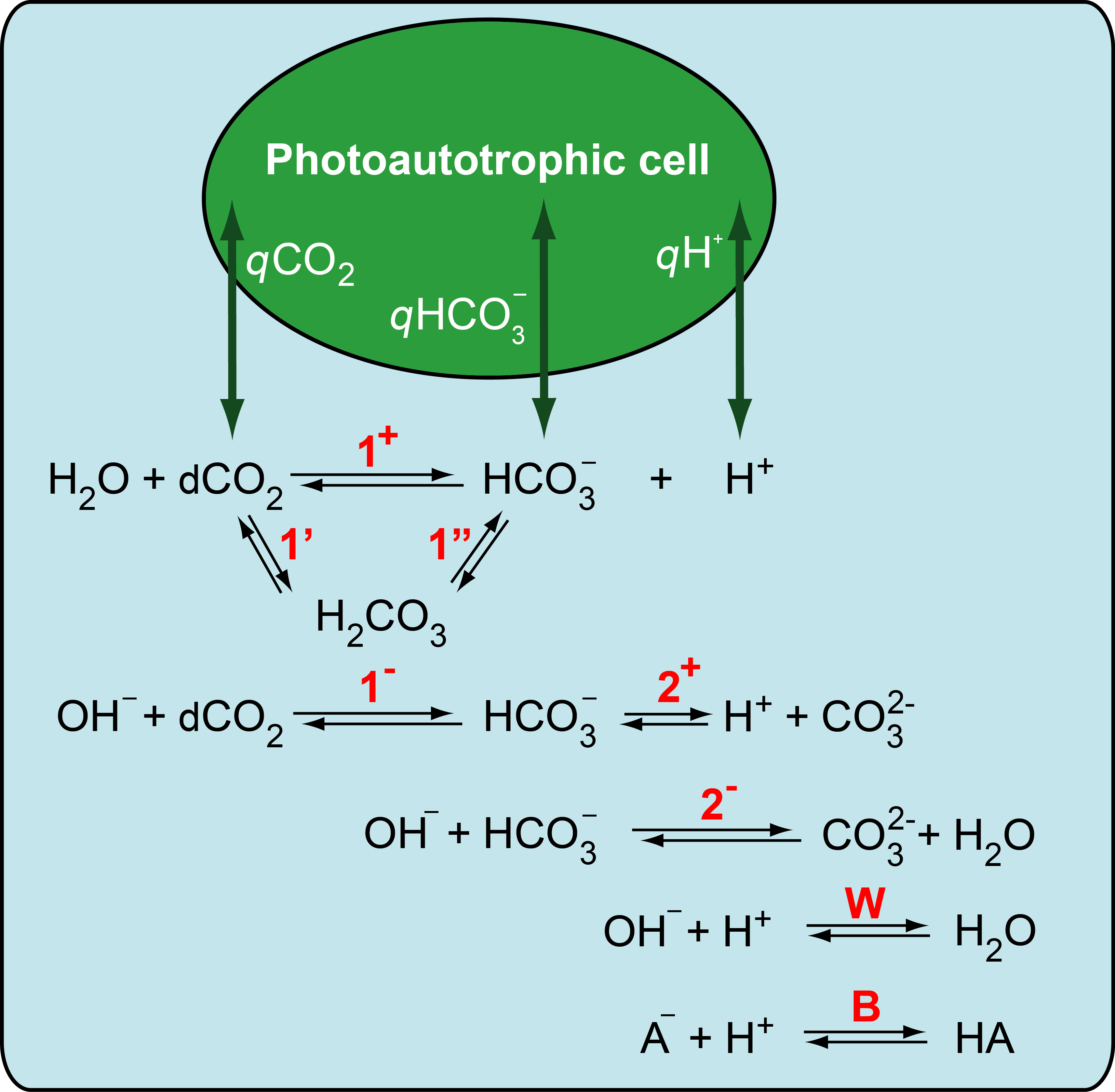
Complete version of the model will be available soon on newly rebuilt version of the webpage.
Model detail
Towards a quantitative assessment of inorganic carbon cycling in photosynthetic microorganisms

Complete version of the model will be available soon on newly rebuilt version of the webpage.
Model detail
M. Trojak, D. Safranek, J. Hrabec, J. Salagovic, F. Romanovska, J. Cerveny: E-Cyanobacterium.org: A Web-Based Platform for Systems Biology of Cyanobacteria. In: Computational Methods in Systems Biology, CMSB 2016, Vol. 9859 of LNCS, pp. 316-322. Springer, 2016. DOI
